Continuous Ingestion
There are currently 13 datasets processed and ingested continuously. For details on the project, see Jira ticket 688 (https://simonscmap.atlassian.net/browse/CMAP-688)
All near real time (NRT) datasets are only ingested to the cluster. Note that dataset replication can be done across any of our servers. See Jira ticket 582 for details on the distributed dataset project (https://simonscmap.atlassian.net/browse/CMAP-582). The following datasets are downloaded, processed, and ingested utilizing run_cont_ingestion.py. This can run via the terminal:
cd ~/Documents/CMAP/cmapdata
python run_cont_ingestion.py
Dataset |
Target Servers |
New Data Available |
|---|---|---|
Sattelite SST |
Rainier, Mariana, Rossby, Cluster |
Daily |
Satellite SSS |
Rainier, Mariana, Rossby, Cluster |
Daily, ~2 week lag |
Satellite CHL |
Mariana, Rossby, Cluster |
~Weekly, ~2 month lag |
Satellite CHL NRT |
Cluster |
~Weekly |
Satellite POC |
Mariana, Rossby, Cluster |
~Weekly, ~2 month lag |
Satellite POC NRT |
Cluster |
~Weekly |
Satellite AOD |
Mariana, Rossby, Cluster |
Monthly |
Satellite Altimetry NRT (Signal) |
Cluster |
Daily |
Satellite Altimetry (Signal) |
Mariana, Rossby, Cluster |
~4x a year |
Satellite PAR (Daily) |
Cluster |
Daily, ~1 month lag |
Satellite PAR NRT (Daily) |
Cluster |
Daily |
Each dataset has a collect and process script.
Collection Scripts
Collection scripts can be found in cmapdata/collect/model and in cmapdata/collect/sat within the dataingest branch of the GitHub repository (https://github.com/simonscmap/cmapdata). Each dataset (with the exception of Argo) adds an entry per file downloaded to tblProcess_Queue.
tblProcess_Queue contains information for each file downloaded for continuous ingestion. It holds the original naming convention for each file, the file’s relative path in the vault, the table name it will be ingested to, the date and time it was downloaded and processed, and a column to take note of download errors.
ID
Original_Name
Path
Table_Name
Downloaded
Processed
Error_Str
Each collection script does the following:
Retries download for any file previously attempted that includes and error message
Get the date of the last succesful download
Attempts to download the next available date range. Start and end dates are specific to each dataset’s temporal resolution and new data availability (see New Data Available column in table above)
Entries for each date a download is attempted for are writted to tblProcess_Queue, with successful downloads denoted by a NULL Error_Str
Dataset-specific logic:
tblPisces_Forecast_cl1 updates the data on a rolling schedule. Each week new data is available, and ~2 weeks of previous data is overwritten by the data producer. GetCMEMS_NRT_MERCATOR_PISCES_continuous.py will re-download these files, and if successfully downloaded, will delete the existing data for that day from the cluster. Simply creating new parquet files and pushing to S3 does not update the data in the cluster, so it is necessary to delete the data first.
The NRT datasets should not overlap with the matching REP datasets. For example, when new data is successfully ingested for tblModis_PAR_cl1, data for that date is deleted from tblModis_PAR_NRT.
A successful download for one day of tblWind_NRT_hourly data results in 24 files. An additional check is in place when finding the last successful date downloaded by including there need to be 24 files for that date. If it is less than 24, it will retry downloading that date.
Data for tblAltimetry_REP_Signal is updated by the data provider sporadically throughout the year. New data is checked weekly. Date range for downloads is based on latest files in the FTP server. See /collect/sat/CMEMS/GetCMEMS_REP_ALT_SIGNAL_continuous.py for details.
If a file download is attempted but fails because no data is available yet, the Original_Name will be the date where data wasn’t available (ex. “2023_08_09”) and the Error_Str will be populated. Each collection script has a retryError function that queries dbo.tblProcess_Queue for any entries where Error_Str is not null. If a file is successfully downloaded on a retry, tblProcess_Queue will be updated with the original file name and date of successful download.
Process Scripts
Each process script does the following:
Pulls newly downloaded files from tblProcess_Queue where Error_Str is NULL.
Does a schema check on the new downloaded file against the oldest NetCDF in the vault
Processes file and adds climatology fields (year, month, week, dayofyear)
Does a schema check on the processed file against the oldest parquet file in the vault
Ingests to on-prem servers (see Target Servers column in table above)
Copies parquet file to S3 bucket for ingestion to the cluster
Updates Processed column in tblProcess_Queue
Adds new entry to tblIngestion_Queue
tblIngestion_Queue contains information for each file processed for continuous ingestion. It holds the file’s relative path in the vault, the table name it will be ingested to, the date and time it was moved to S3 (Staged), and the date and time it was added to the cluster (Started and Ingested).
ID
Path
Table_Name
Staged
Started
Ingested
Once all new files have been processed from tblProcess_Queue and added to tblIngestion_Queue, trigger the ingestion API. The URL is saved in ingest/credentials.py as S3_ingest. It is best to only trigger the ingestion API once, which is why the snippet below is run after files for all datasets have been processed. See Jira ticket 688 for additional details: (https://simonscmap.atlassian.net/browse/CMAP-688)
requests.get(cr.S3_ingest)
After all files have successfully ingested to the cluster (Ingested will be filled with the date and time it was completed), each dataset will need updates to tblDataset_Stats. In run_cont_ingestion.py, updateCIStats(tbl) formats the min and max times to ensure the download subsetting and viz page works properly. In short, time must be included, along with the ‘.000Z’ suffix.
Troubleshooting
Occasionally datasets will have days missing, resulting in a date being retried on each new run of run_cont_ingestion.py. In some cases, data will never be provided for these dates. This information can be found in the documentation provided by each data provider. For example, the SMAP instrument used for SSS data experienced downtime between Aug 6 - Sept 23 2022 (see Missing Data section: https://remss.com/missions/smap/salinity/). That date range was deleted from tblProcess_Queue to prevent those dates from being rechecked each run.
If there are known issues of data already ingested that the data producer has fixed, the entry for the impacted dates should be deleted from tblProcess_Queue and tblIngestion_Queue and redownloaded. Data should be delete from impacted on-prem servers and the cluster as applicable before reingestion.
Each dataset’s processing script has checks for changes in schema. Some data providers will change the dataset name when a new version is processed, but not all. If a processing script finds a schema change for a dataset that has the same name / ID / version number, a new dataset should be made in CMAP with a suffix denoting a new change log iteration. For example, tblModis_CHL_cl1 is a reprocessed version of tblModis_CHL.
Batch Ingestion
The following datasets are to be ingested monthly. Due to the nature of updates done by the data provider, each month of Argo is a new dataset. These datasets will be ingested via batch ingestion instead of appending to existing tables like the datasets described above. See the outside large dataset walkthrough for details on Argo processing.
Dataset |
Target Servers |
New Data Available |
|---|---|---|
Argo REP Core |
Cluster |
Monthly |
Argo REP BGC |
Cluster |
Monthly |
Continuous Ingestion Badge on Website
The CMAP Catalog page includes a filter for Continuously Updated datasets and displays badges for each applicable dataset.
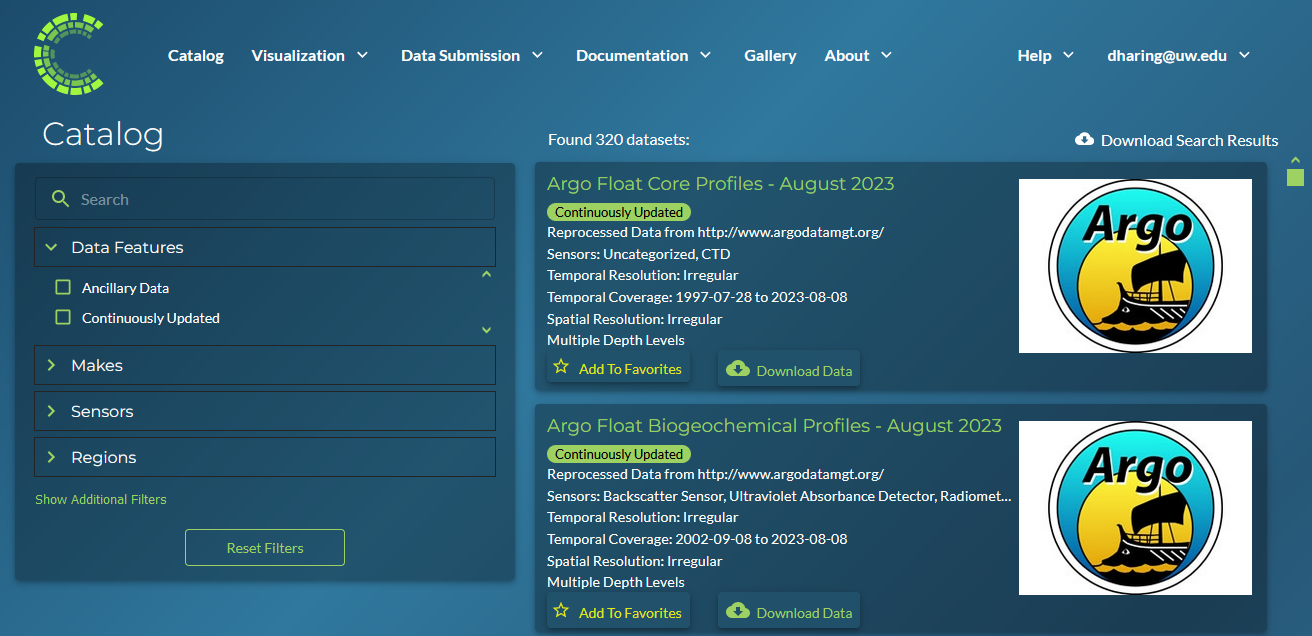
The badges and filter call the uspDatasetBadges stored procedure, which in turn calls the udfDatasetBadges() function. As Argo datasets are a batch ingestion, they are not included in tblProcess_Queue. In order to have the badges display for Argo datasets, a union was done for any Argo REP table, regardless of month.
select distinct table_name, ci = 1 from tblProcess_Queue
union all
select distinct table_name, ci = 1 from tblvariables where Table_Name like 'tblArgo%_REP_%'
Sea Surface Salinity Walkthrough
There are two version of Sea Surface Salinity (SSS) data. For details on the differences see Jira ticket 754 (https://simonscmap.atlassian.net/browse/CMAP-754). Continuous ingestion downloads the REMSS SMAP data. The previous version of SSS data in CMAP was collected from V4.0 provided by REMSS. The updates in the V5.0 release recalculated historic data (details can be found here: https://remss.com/missions/smap/salinity/), which meant we could no longer append new data to the existing table.
Due to the release of V5.0, a new table was made with a “change log” suffix of 1. New SSS data is currently ingested into tblSSS_NRT_cl1. The tblSSS_NRT table can be retired and removed from the databases after users have been notified via a news update on the homepage. A typical wait time has been one month after publishing a news story before a dataset can be removed.
Download SSS Data
The data is downloaded using wget, calling the data.remss URL with the year and day of year of the data:
file_url = f'https://data.remss.com/smap/SSS/V05.0/FINAL/L3/8day_running/{yr}/RSS_smap_SSS_L3_8day_running_{yr}_{dayn_str}_FNL_V05.0.nc'
As with each continuously ingested dataset, there is a function to retry any previous dates that resulted in an error. Errors are generated by either a successful, but empty download, or a failed download attempt.
The first function run in the GetREMSS_SSS_cl1_continuous.py script is:
def retryError(tbl):
qry = f"SELECT Original_Name from dbo.tblProcess_Queue WHERE Table_Name = '{tbl}' AND Error_Str IS NOT NULL"
df_err = DB.dbRead(qry, 'Rainier')
dt_list = df_err['Original_Name'].to_list()
if len(dt_list)>0:
dt_list = [dt.datetime.strptime(x.strip(), '%Y_%m_%d').date() for x in dt_list]
for date in dt_list:
get_SSS(date, True)
This function checks tblProcess_Queue for any previous errors and runs a retry on the download.
The get_SSS function formats the date into the necessary day of year format and attempts a download via wget. If the download is a retry, the entry for that date in tblProcess_Queue will be updated with the original file name and the date of the successful download. If the download is not a retry, a new entry will be added to tblProcess_Queue. If any failure occurs, the error_str will be populated and the original file name will be populated with the date of the data that failed to download (ie “2023_08_26”). This string is converted to a date for future retries.
def get_SSS(date, retry=False):
yr = date.year
dayn = format(date, "%j")
dayn_str = dayn.zfill(3)
file_url = f'https://data.remss.com/smap/SSS/V05.0/FINAL/L3/8day_running/{yr}/RSS_smap_SSS_L3_8day_running_{yr}_{dayn_str}_FNL_V05.0.nc'
save_path = f'{vs.satellite + tbl}/raw/RSS_smap_SSS_L3_8day_running_{yr}_{dayn_str}_FNL_V05.0.nc'
wget_str = f'wget --no-check-certificate "{file_url}" -O "{save_path}"'
try:
os.system(wget_str)
Error_Date = date.strftime('%Y_%m_%d')
Original_Name = f'RSS_smap_SSS_L3_8day_running_{yr}_{dayn_str}_FNL_V05.0.nc'
## Remove empty downloads
if os.path.getsize(save_path) == 0:
print(f'empty download for {Error_Date}')
if not retry:
metadata.tblProcess_Queue_Download_Insert(Error_Date, tbl, 'Opedia', 'Rainier','Download Error')
os.remove(save_path)
else:
if retry:
metadata.tblProcess_Queue_Download_Error_Update(Error_Date, Original_Name, tbl, 'Opedia', 'Rainier')
print(f"Successful retry for {Error_Date}")
else:
metadata.tblProcess_Queue_Download_Insert(Original_Name, tbl, 'Opedia', 'Rainier')
except:
print("No file found for date: " + Error_Date )
metadata.tblProcess_Queue_Download_Insert(Error_Date, tbl, 'Opedia', 'Rainier','No data')
After the retry function is run, the last date that was successfully downloaded is retrieved by checking tblIngestion_Queue, tblProcess_Queue, and max date from the cluster.
def getMaxDate(tbl):
## Check tblIngestion_Queue for downloaded but not ingested
qry = f"SELECT Path from dbo.tblIngestion_Queue WHERE Table_Name = '{tbl}' AND Ingested IS NULL"
df_ing = DB.dbRead(qry, 'Rainier')
if len(df_ing) == 0:
qry = f"SELECT max(path) mx from dbo.tblIngestion_Queue WHERE Table_Name = '{tbl}' AND Ingested IS NOT NULL"
mx_path = DB.dbRead(qry,'Rainier')
path_date = mx_path['mx'][0].split('.parquet')[0].rsplit(tbl+'_',1)[1]
yr, mo, day = path_date.split('_')
max_path_date = dt.date(int(yr),int(mo),int(day))
qry = f"SELECT max(original_name) mx from dbo.tblProcess_Queue WHERE Table_Name = '{tbl}' AND Error_str IS NOT NULL"
mx_name = DB.dbRead(qry,'Rainier')
if mx_name['mx'][0] == None:
max_name_date = dt.date(1900,1,1)
else:
yr, mo, day = mx_name['mx'][0].strip().split('_')
max_name_date = dt.date(int(yr),int(mo),int(day))
max_data_date = api.maxDateCluster(tbl)
max_date = max([max_path_date,max_name_date,max_data_date])
else:
last_path = df_ing['Path'].max()
path_date = last_path.split('.parquet')[0].rsplit(tbl+'_',1)[1]
yr, mo, day = path_date.split('_')
max_date = dt.date(int(yr),int(mo),int(day))
return max_date
The date range to check data for is specific to each dataset depending on the temporal scale and typical delay in new data availability from the data producer. For SSS data from REMSS, there is a NetCDF file for each day (timedelta(days=1)), and new data is generally available on a two week delay.
Note
If new data has not been published by REMSS for a month or so, emailing their support account (support@remss.com) has been helpful to restart their processing job.
Process SSS Data
The first step of process_REMSS_SSS_cl1_continuous.py is to pull the list of all newly downloaded files. The tblProcess_Queue and tblIngestion_Queue tables only live on Rainier, so that server needs to be specified when retrieving the new files ready for processing:
qry = f"SELECT Original_Name from tblProcess_Queue WHERE Table_Name = '{tbl}' AND Path IS NULL AND Error_Str IS NULL"
flist_imp = DB.dbRead(qry,'Rainier')
flist = flist_imp['Original_Name'].str.strip().to_list()
The schema of the newly downloaded NetCDF is compared against the oldest NetCDF in the vault. If any new columns are added or renamed, the processing will exit and the data will not be ingested. After the NetCDF has gone through all the processing steps, the schema of the finalized parquet file is checked against the oldest parquet file in the vault. Again, if there are any differences the processing will exit and the data will not be ingested. This logic is present in all continuously ingested (CI) datasets. Additional steps done for all CI datasets include: adding climatology columns, updating tblProcessQueue with processing datetime, saving parquet file to vault, pushing parquet from vault to S3 bucket, and adding a new entry to tblIngestion_Queue.
Processing logic specific to SSS includes: pulling time from NetCDF coordinate, extracting single variable from NetCDF (sss_smap), and mapping longitude from 0, 360 to -180, 180. Because SSS data is frequently accessed, it is ingested into all on-prem servers, as well as the cluster.
A single parquet file is ingested into on-prem servers simultaneously using Pool. The current BCP wrapper creates a temporary csv file with the table name and server name in it, to allow for multiple ingestions at once. The multiprocessing is not done on multiple files for the same dataset across servers as the current file naming convention could cause clashes if overwritten.
The list of original file names is looped through for processing:
for fil in tqdm(flist):
x = xr.open_dataset(base_folder+fil)
df_keys = list(x.keys())
df_dims = list(x.dims)
if df_keys != test_keys or df_dims!= test_dims:
print(f"Check columns in {fil}. New: {df.columns.to_list()}, Old: {list(x.keys())}")
sys.exit()
x_time = x.time.values[0]
x = x['sss_smap']
df_raw = x.to_dataframe().reset_index()
df_raw['time'] = x_time
x.close()
df = dc.add_day_week_month_year_clim(df_raw)
df = df[['time','lat','lon','sss_smap','year','month','week','dayofyear']]
df = df.sort_values(["time", "lat","lon"], ascending = (True, True,True))
df = dc.mapTo180180(df)
if df.dtypes.to_dict() != test_dtype:
print(f"Check data types in {fil}. New: {df.columns.to_list()}")
sys.exit()
fil_name = os.path.basename(fil)
fil_date = df['time'][0].strftime("%Y_%m_%d")
path = f"{nrt_folder.split('vault/')[1]}{tbl}_{fil_date}.parquet"
df.to_parquet(f"{nrt_folder}{tbl}_{fil_date}.parquet", index=False)
metadata.tblProcess_Queue_Process_Update(fil_name, path, tbl, 'Opedia', 'Rainier')
s3_str = f"aws s3 cp {tbl}_{fil_date}.parquet s3://cmap-vault/observation/remote/satellite/{tbl}/nrt/"
os.system(s3_str)
metadata.tblIngestion_Queue_Staged_Update(path, tbl, 'Opedia', 'Rainier')
a = [df,df,df]
b = [tbl,tbl,tbl]
c = ['mariana','rossby','rainier']
with Pool(processes=3) as pool:
result = pool.starmap(DB.toSQLbcp_wrapper, zip(a,b,c))
pool.close()
pool.join()